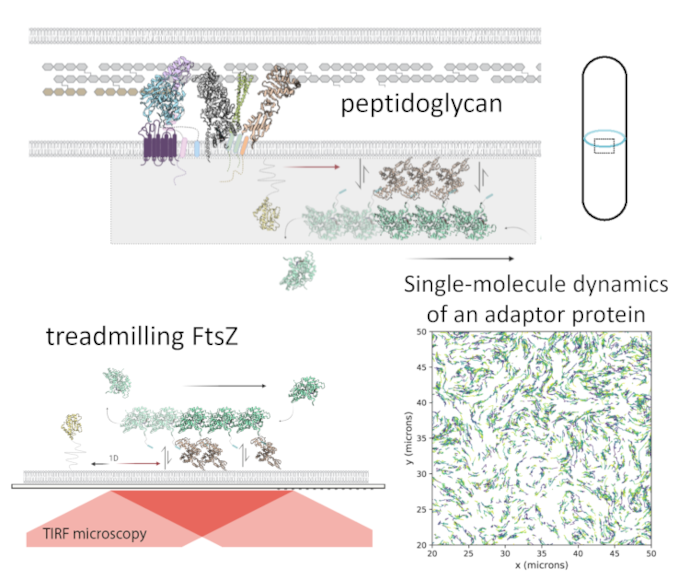
The breakthrough in vivo studies in the field of bacterial cell biology revealed that treadmilling cytoskeleton protein FtsZ could actively move transmembrane peptidoglycan synthases, and such dynamic coupling is critical for bacterial division. However, the underlying molecular mechanisms behind the coordination is still incomplete, even for a model organism as E.coli. In our previous work we reconstituted a part of bacterial division machinery from purified components and found that membrane-bound proteins can self-organize with the FtsZ filaments into chiral rotating rings. By employing single-molecule tracking and advanced image analysis, we will aim to understand the underlying mechanisms behind the transport of peptidoglycan synthesis machinery by FtsZ cytoskeleton in a tunable reconstitution assay.

Related publications:
N. Baranova, P. Radler, V. Hernandez-Rocamora, C. Alfonso, M. Lopez Pelegrin, G. Rivas, W. Vollmer and M. Loose (2020) FtsZ assembles the divisome by a diffusion-and-capture mechanism. Nature Microbiology, doi.org/10.1038/s41564-019-0657-5
P. Radler*, N. Baranova*, P. Caldas, M. Lopez Pelegrin, M. Loose (2022). In vitro reconstitution of Escherichia coli divisome activation. Nature Communications 13(1). doi: 10.1038/S41467-022-30301-Y *- equal contribution